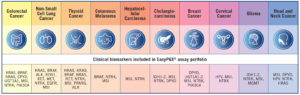
EasyPGX®: the dry qPCR revolution against cancer
EasyPGX® is the complete RT-qPCR in vitro diagnostic solution with the most comprehensive portfolio of assays for molecular oncology.
With its revolutionary ready-to-use, dry-format, pre-aliquoted, 8-well reaction strips, EasyPGX® is suited to use in any laboratory setting, assuring rapid, high-performance, standardised results with easy handling.
EasyPGX® product line – key features
- Ready to use: ready-to-use dry reagents,pre-aliquoted in 8-well RT-PCR strips.
- Easy to use: fewer pipetting steps needed for reaction set up with a total HoT <10 min. No need for freezing-thawing cycles or pipetting on ice.
- High sensitivity: limit of detection as low as 0.5%.
- Flexible sample requirement: validated for low quantity and low quality DNA, ct DNA and RNA from different input materials such as FFPE and liquid biopsies.
- Turnaround time: from tissue to result in less than 3 hours with only 10 minutes of hands-on time.
- Fast: from sample to result in under 3 hours.
- Flexible: runs multiple targets in one experiment.
- Automatic data analysis: includes dedicated automated data analysis and interpretation software.
- Room temperature shipping and storage: complete master mix in a dry format that is stable at room temperature.
- Standards included: includes positive and negative controls for the validation of each session.
- Regulatory: Compliant with the current EU IVDR regulation (2017/746).
EasyPGX system workflow: from tissue to result in less than 3 hours.

Main features
Detection of the main mutations of exon 2 (codons 12, 13), of exon 3 (codons 59, 61) and of exon 4 (codons 117, 146) of the gene KRAS using 8
oligo mixes. Each mix allows the co-amplification of one or more mutated alleles plus an endogenous control gene. A specific oligo control mix enables
the evaluation of the quality and the quantity of the DNA in each sample.
Controls
The kit is provided with the following controls:
- Positive control sample containing a mixture of synthetic DNA sequences that correspond to each mutation detected by this kit in a background of wild-type genomic DNA;
- Negative control.
Sensitivity
The kit allows the detection of low percentages of mutated allele in presence of high amounts of wild-type genomic DNA by real-time amplification with sequence-specific probes marked with FAMb and HEX. LOD down to 0.5%.
Starting material
The kit allows the analysis of DNA extracted from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissues and plasma*.
*Please note that extraction from plasma is sold separately (cat.no. H8040)
Main features
Detection of the main mutations of codon 600 of the gene BRAF using 4 oligo mixes. Each mix allows the co-amplification of one or more mutated alleles plus an endogenous control gene. A specific oligo control mix enables the evaluation of the quality and the quantity of the DNA in each sample.
Controls
The kit is provided with the following controls:
- Positive control sample containing a mixture of synthetic DNA sequences that correspond to each mutation detected by this kit in a background of wild-type genomic DNA;
- Negative control.
Sensitivity
The kit allows the detection of low percentages of mutated allele in presence of high amounts of wild-type genomic DNA by real-time amplification with sequence-specific probes marked with FAMb and HEX. LOD down to 0.5%.
Starting material
The kit allows the analysis of DNA extracted from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissues and plasma*.
*Please note that extraction from plasma is sold separately (cat.no. H8040)
Main features
Detection of the main mutations of exons 18, 19, 20, 21 of EGFR gene using 8 oligo mixes. Each mix allows the co-amplification of one or more mutated alleles plus an endogenous control gene. A specific oligo control mix enables the evaluation of the quality and the quantity of the DNA in each sample.
Controls
The kit is provided with the following controls:
- Positive control sample containing a mixture of synthetic DNA sequences that correspond to each mutation detected by this kit in a background of wild-type genomic DNA;
- Negative control.
Sensitivity
The kit allows the detection of low percentages of mutated allele in presence of high amounts of wild-type genomic DNA by real-time amplification with sequence-specific probes marked with FAMb and HEX. LOD down to 0.5%.
Starting material
The kit allows the analysis of DNA extracted from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissues and plasma*.
*Please note that extraction from plasma is sold separately (cat.no. H8040)
Main features
Detection of the main mutations of exon 2 ( codons 12, 13), of exon 3 (codons 59, 61) and of exon 4 (codons 117, 146) of NRAS gene using 8 oligo mixes. Each mix allows the co-amplification of one or more mutated alleles plus an endogenous control gene. A specific oligo control mix enables the evaluation of the quality and the quantity of the DNA in each sample.
Controls
The kit is provided with the following controls:
- Positive control sample containing a mixture of synthetic DNA sequences that correspond to each mutation detected by this kit in a background of wild-type genomic DNA;
- Negative control.
Sensitivity
The kit allows the detection of low percentages of mutated allele in presence of high amounts of wild-type genomic DNA by real-time amplification with sequence-specific probes marked with FAMb and HEX. LOD down to 0.5%.
Starting material
The kit allows the analysis of DNA extracted from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissues and plasma*.
*Please note that extraction from plasma is sold separately (cat.no. H8040)
Main features
Detection of the main chromosomal translocations involving ALK, ROS1, RET and the MET exon 14 skipping. Each mix allows the co-amplification of one or more fusions plus an endogenous control gene.
Controls
- Positive control containing a mixture of synthetic RNA-DNA sequences that correspond to the main fusions detected by the kit;
- Negative control.
Starting material
RNA from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissues and cytological samples.
Main features
Detection, by allelic discrimination, of the DPYD gene polymorphisms DPYD*2A (IVS14+1G>A,c.1905+1G>A,rs3918290), DPYD*13 (c.1679T>G,rs55886062), DPYD D949V (c.2846A>T, rs67376798), DPYD*6 (c.2194G>A, rs1801160) and DPYD IVS10 (c.1129–5923C>G, rs75017182),associated with the toxicity due to the treatment with Fluoropyrimidines, using 4 oligo mixes. Each mix allows the co-amplification of the mutant sequence (FAM) as well as the wild-type sequence (HEX).
Controls
The kit is provided with the following controls:
- DPYD WT positive control: Positive control DNA containing a mixture of synthetic wild-type DNA sequences for the DPYD polymorphisms analyzed;
- DPYD MT positive control: Positive control DNA containing a mixture of synthetic mutant DNA sequences for the DPYD polymorphisms analyzed;
- Negative control.
Starting material
The kit allows the analysis of genomic DNA extracted from whole blood.
Main features
Detection, by allelic discrimination, of the UGT1A1 gene polymorphisms UGT1A1*1 (TA)6, UGT1A1*28 (TA)7, UGT1A1*36 (TA)5 and UGT1A1*37 (TA)8, associated with the toxicity due to the treatment with Irinotecan, using 1 oligo mix. UGT1A1 mix contains HEX labeled probes for UGT1A1*28 and UGT1A1*37 and FAM labeled probes for UGT1A1*1 and UGT1A1*36.
Controls
The kit is provided with the following controls:
- UGT1A1 WT positive control: Positive control DNA containing synthetic wild-type UGT1A1*1/*1 DNA sequence;
- UGT1A1 MT positive control: Positive control DNA containing a synthetic mutant UGT1A1*28/*28 DNA sequence;
- Negative control.
Starting material
The kit allows the analysis of genomic DNA extracted from whole blood.
Main features
Detection of the main mutations of exon 2 (codons 12,13), of exon 3 (codons 61) of the genes KRAS, NRAS, HRAS and of the codons 600 and 601 of the gene BRAF using 8 oligo mixes. Each mix allows the co-amplification of one or more mutated alleles plus an endogenous control gene.
Controls
The kit is provided with the following controls:
- Positive control sample containing a mixture of synthetic DNA sequences that correspond to each mutation detected by this kit in a background of wild-type genomic DNA;
- Negative control.
Sensitivity
The kit allows the detection of low percentages of mutated allele in presence of high amounts of wild-type genomic DNA by real-time amplification
with sequence-specific probes marked with FAM and HEX. LOD down to 0.5%.
Starting material
The kit allows the analysis of DNA extracted from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissues, plasma*, and cytological samples.
*Please note that extraction from plasma is sold separately (cat.no. H8040)
Main features
Quantitative determination of DNA fragments with dimensions > 200bpThe quantitative analysis of DNA extracted from the faeces in association with FOBT test, represents a useful tool to improve early diagnosis of CRC allowing to estimate the probability of the CRC presence.
Controls
- Positive control, genomic DNA at known concentration;
- Standard, genomic DNA at known concentration to diluite for creating the standard curve;
- Spike, control DNA to add into the samples to verify the inhibition;
- Negative control.
Starting material
DNA from faecal specimens.
Main features
Detection of T790M and C797S (c.2389 T>A, c.2390 G>C) of the EGFR gene.
Each mix allows the co-amplification of one or more mutated alleles plus an endogenous control gene.
Controls
- Positive control sample containing a mixture of synthetic DNA sequences that correspond to each mutation detected by this kit in a background of wild-type genomic DNA;
- Negative control.
Starting material
DNA from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissues and plasma*.
*Please note that extraction from plasma is sold separately (cat.no. H8040)
Main features
Detection of the main mutations of IDH1 gene (codons 105 and 132) and IDH2 gene (codons 140 and 172). Each mix allows the co-amplification of one or more mutated alleles plus an endogenous control gene. A specific oligo control mix enables the evaluation of the quality and the quantity of the DNA in each sample.
Controls
- Positive control sample containing a mixture of synthetic DNA sequences that correspond to each mutation detected by this kit in a background of wild-type genomic DNA;
- Negative control.
Starting material
DNA from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissues and plasma*.
*Please note that extraction from plasma is sold separately (cat. n. H8040).
Main features
Detection of the chromosomal translocations involving RET/PTC1: CCDC6-RET; RET/PTC2: PRKAR1A-RET; RET/PTC3: NCOA4-RET and PAX8/PPARG. Each mix allows the co-amplification of one or more fusions plus an endogenous control gene.
Controls
- Positive control sample containing a mixture of synthetic RNA-DNA sequences that correspond to the main fusions detected by the kit;
- Negative control.
Starting material
RNA from fresh, frozen, formalin-fixed paraffinembedded (FFPE) tissues and cytological samples.
Main features
Detection of 8 mononucleotide “quasi – monomorphic” markers: BAT-25, BAT-26, NR-21, NR-22, NR-24, NR-27, CAT-25 and MONO-27 by Real Time PCR and subsequent analysis of the targets based on the denaturation profile. The test allows, accurately and with reduced “hands-on time”, to detect the microsatellite instability in tumor samples.
Controls
- Positive control, genomic DNA corresponding to a stable profile;
- Negative control.
Starting material
DNA from fresh, frozen, formalin fixed paraffin embedded (FFPE) tissue. Comparison with normal tissue or blood is not necessary for the analysis of results.
Main features
Identification of 14 High Risk genotypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) of Human Papilloma Virus (HPV) by amplifying the E6 and E7 oncogenes.
Each mix allows the co-amplification of the genotype-specific HPV targets plus an endogenous control gene.
Starting material
DNA from cervical swabs and formalin-fixed paraffin-embedded (FFPE) tissue.
Main features
Detection of the main fusion variants of the NTRK1, NTRK2 and NTRK3 genes by one step Real Time PCR. Each mix allows the co-amplification of one or more fusions plus an endogenous control gene.
Controls
- Control assay: an expressed gene is amplified in the channel stably and independently from the original tumor tissue (Pan-cancer housekeeping gene) in order to verify the correct execution of the amplification process, the RNA quantity used and the possible presence of inhibitors that can cause false negative results;
- Positive control: the test contains a mix of RNA-DNA synthetic sequences which correspond with the main fusions detected by the kit;
- Negative control.
Starting material
RNA from fresh, frozen, formalin-fixed paraffinembedded (FFPE) tissues and cytological samples.
Main features
Detection of the main mutations of codons 345, 420, 542, 545, 546, 1047, 1049 of the PIK3CA gene.
Each mix allows the co-amplification of one or more mutated alleles plus an endogenous control gene.
Controls
- Control assay: there is a specific assay to evaluate the DNA content of the sample. In addition, in every mix a detected endogenous control gene allows to verify the correct execution of the amplification process and the possible presence of inhibitors that can cause false negative results;
- Positive control: contains a mix of synthetic DNA which corresponds to the mutations detected by the kit and the genomic wild-type DNA;
- Negative control.
Starting material
The kit allows the analysis of DNA extracted from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissues and plasma*.
*Please note that extraction from plasma is sold separately (cat.no. H8040)
Main features
Qualitative detection, by RT-PCR and subsequent analysis of the targets based on the denaturation profile, of the methylation status of 12
CpG sites located in the promoter of the MGMT gene. The kit includes reagents for sodium bisulfite treatment of DNA extracted before
methylation analysis, which converts unmethylated cytosines to uracil.
Controls
- Methylated positive control.
- Unmethylated positive control.
- Negative control.
Starting material
The kit allows the analysis of DNA extracted from formalin-fixed paraffin-embedded (FFPE) tissues.
Main features
Detection of the most frequent FGFR3 gene somatic mutations in exons 7, 9, 14 and FGFR2, FGFR3 gene fusions using 8 oligo mixes. Each mix allows the co-amplification the target plus an endogenous control gene. A specific oligo control mix enables the evaluation of the quality and the quantity of each sample.
Controls:
- Positive control sample containing a mixture of synthetic DNA sequences positive for all detectable targets, in a background of wild-type DNA and RNA.
- Negative control
Sensitivity:
The kit demonstrated a specificity of 100% and a sensitivity >97%.
Starting Material:
The kit allows the analysis of DNA and RNA extracted from fresh, frozen, formalin-fixed paraffin-embedded (FFPE) tissue.

Enter your information below to download the brochures
Category
Realtime PCR


