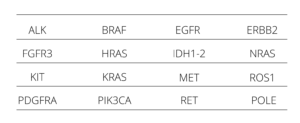
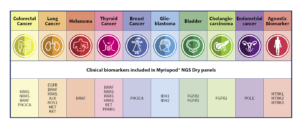
Myriapod NGS Dry line includes NGS panels for DNA and RNA sequencing dedicated to the study of predictive- diagnostic SNV (SNP, In/del, fusions) clinically relevant in the most frequent cancer types as: colon, lung, melanoma, central nervous system, GIST, thyroid, and bladder, and also independent tissue biomarkers, like NTRK 1, 2 and 3 gene fusions.
Main features:
- Ready to use: Dry, ready-to-use, in pre-aliquoted strip reagent format
- Simple: the easiest and fastest workflow on the market for NGS libraries preparation, with a hands-on time < 2 hours and a turnaround time, from sample to sequencing, of 8 hours.
- Flexible: optimized on Illumina® Miseq™, MiniSeq™ and iSeql100™ with the flexibility to test from 2 to 46 samples/run.
- Low amount and quality of input: validated to work from 10 ng/reactions of DNA and RNA extracted from FFPE or cfDNA.
- Certified: fully CE-IVD protocol, from sample to result with data processing through Myriapod® NGS Data Analysis Software, fully automatic, bioinformatic analysis local system without any needs to send out data or use cloud based solutions.
Myriapod® NGS data Analysis: the CE-IVD solution for local NGS data analysis
Myriapod® NGS Data Analysis is a complete, in vitro diagnostic and easy-to-use bioinformatics solution, which come with the Myriapod® NGS Data Analysis Software and its own dedicated workstation. Myriapod NGS solution provides simplicity in the analysis for fast and easy reporting. Further, the data management system offers the highest level of security with a completely localized data analysis, so sensitive data is not sent over the network or to a cloud system. Myriapod NGS Data Analysis solution complies with all the EU General Data Protection Regulations (GDPR).
Enter your information below to download the brochure
Outsourcing of data and sample analysis do not eliminate the legal responsibility of the laboratory in the processing of personal data, even if protected by complex contracts subscribed with the service provider and even if the last one is certified according to a standard for “Information Security Management” (eg. ISO 27001).
Learn more by downloading the flyer
Category
Next Generation Sequencing